Finding the "Goods" in a Molecular Jungle
Finding the "Goods" in a Molecular Jungle
The overarching problems that we try to address are 1) how do we define the molecular signatures of a biological system in response to stress caused by the environment, chronic diseases and aging? and 2) how can we manipulate the stress response to do good to the biological system?
For this purpose we combine chemistry, separation technologies and mass spectrometry to detect, identify and quantify biomolecules in complex matrices. We develop analytical methods to explore the metabolome and proteome as comprehensive collections of molecular messengers of cellular response. We have collaborations with clincial research groups in the area of disease biomarkers.
Biomolecular Mass Spectrometry
We develop and apply methods based on ion mobility mass spectrometry for the characterization of small molecules and biopolymers. We use ion mobility mass spectrometry as orthogonal method for the analysis of small molecules in complex matrices and biological systems.
Analytical Metabolomics
We develop and apply biomolecular mass spectrometry method for detecting and identifying metabolites and xenobiotics in complex biological matrices. Our methods enable us to determine the response of the metabolome to disease, host-environment interactions, and exposure in order to better define the metabolic pathways and biological networks affected by the perturbation with the goal to discvery biomarkers and guide intervention strategies.
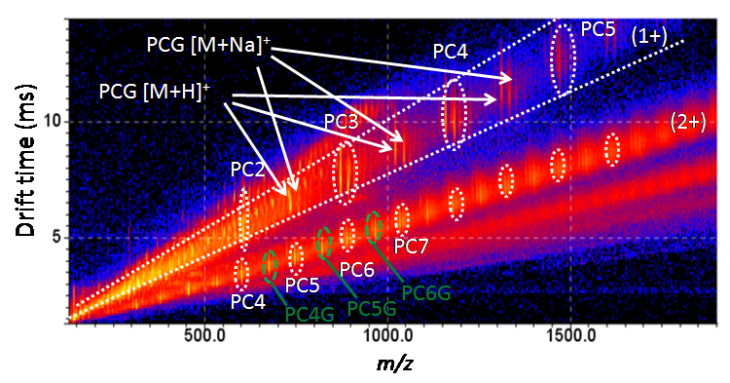
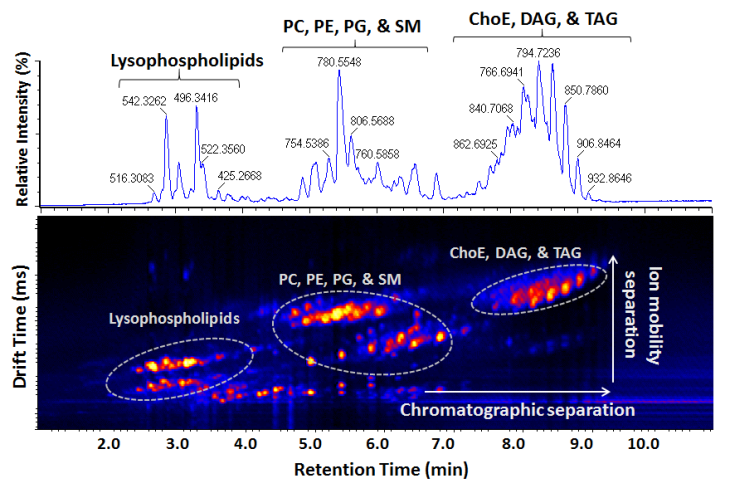
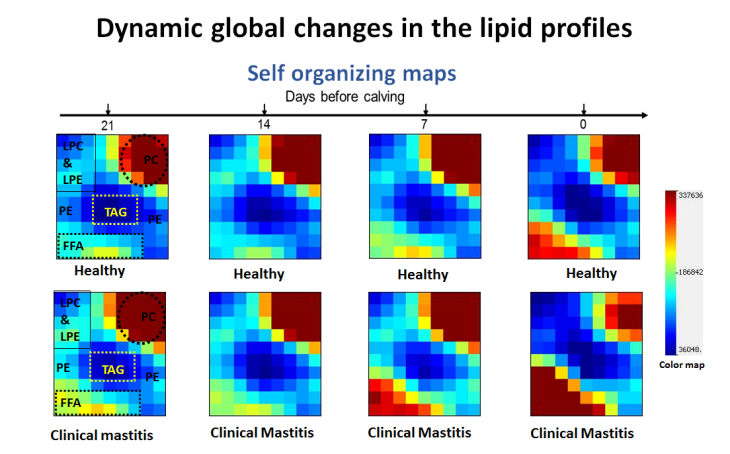
Development and Application of Mass Spectrometry-driven Methods for Detection and Characterization of Biomolecular Signatures of Diseases (Zandkarimi et al. J. Dairy Sci. 2018)
Mass Spectrometry-based Proteomics and PTM mapping
We use a combination of biochemical methods, analytical chemistry and mass-spectrometry-based proteomics to understand dysregulation in disease and environmental exposure. We develop proteomics methods to understand the mechansims of aging, hypoxic stress in cancer and the consequence of radiation.
Ion mobility-enhanced MS(E)-based label-free analysis reveals effects the low-dose radiation post contextual fear conditioning training on the mouse hippocampal proteome. Huang et al. J. Proteomics 2016
Label-free proteomics assisted by affinity enrichment for elucidating the chemical reactivity of the liver mitochondrial proteome toward adduction by the lipid electrophile 4-hydroxy-2-nonenal (HNE). Tzeng & Maier, Front Chem. 2016
Protein modification by electrophilic lipoxidation products; adduct formation, chemical strategies and tandem mass spectrometry for their detectioan and identifcation. Vasil'ev YV, Tzeng SC, Huang L, Maier CS. Mass Spectrometry Rev. 2014

We study the proteome response to changes in redox homeostasis with relevance to environmental exposure, chronic diseases and aging.
For this purpose we develop and advance analytical proteomic techniques for defining the chemical diversity of insult by oixidative and electrophile stress. We use chemoselective methods coupled to mass spectrometry to define the chemical signatures of oxidative stress at the systems level which the goal to use oxidative signatures one day for diagnosis of health status in personalized medicine and therapy monitoring.
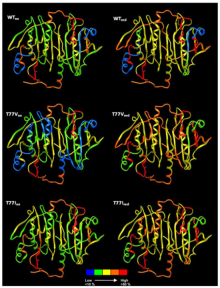
Structural Proteomics
We study how proteins work and interact under conditions mimicking their biological environments. We study protein mobility and interactions by solution-phase hydrogen/deuterium exchange mass spectrometry (HDX-MS).